安装与介绍
# install.packages('tidyverse', repos = "https://mirrors.tuna.tsinghua.edu.cn/CRAN/")
library(tidyverse)
── Attaching packages ──────────────────────────────────── tidyverse 1.3.0 ──
✓ ggplot2 3.3.0 ✓ purrr 0.3.3
✓ tibble 3.0.0 ✓ dplyr 0.8.5
✓ tidyr 1.0.2 ✓ stringr 1.4.0
✓ readr 1.3.1 ✓ forcats 0.5.0
── Conflicts ─────────────────────────────────────── tidyverse_conflicts() ──
x dplyr::filter() masks stats::filter()
x dplyr::lag() masks stats::lag()
管道操作符
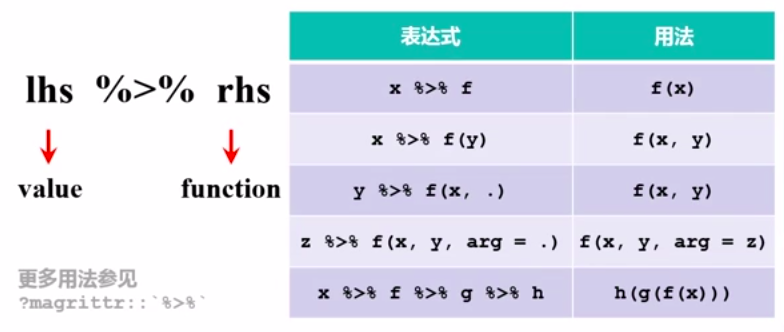
图片alt
cjb <- read.csv("/home/wy/Downloads/cjb.csv",
header = TRUE,
stringsAsFactors = FALSE,
fileEncoding = "UTF-8")
cjb %>% head
head(cjb) # 同上
cjb %>% head(n=3) # cjb 默认为第一个参数
head(cjb,n=3) # 同上
dplyr包使用
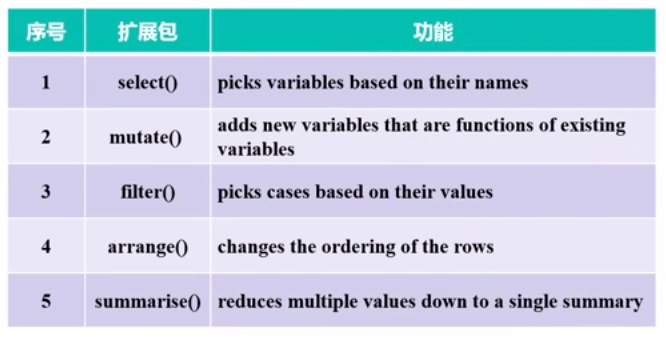
图片alt
选择列
cjb %>%
select(xm,yw,sx) %>%
set_names(c('姓名','语文','数学')) %>%
head(n=3)
# 姓名 语文 数学
# 1 周黎 94 82
# 2 汤海明 87 94
# 3 舒江辉 92 79
cjb %>%
select(1,4:12) %>%
head(n=3)
cjb %>%
select(xm,yw:sw) %>%
head(n=3)
# xm yw sx wy zz ls dl wl hx sw
# 1 周黎 94 82 96 97 97 98 95 94 88
# 2 汤海明 87 94 89 95 94 94 90 90 89
# 3 舒江辉 92 79 86 98 95 96 89 94 87
增加或者修改列
cjb %>%
# 将bj,xb,wlfk转化因子
mutate_at(vars(bj,xb,wlfk),factor) %>%
# 求出.(表示前面对象)的4~12列总和
mutate(zcj = rowSums(.[4:12])) %>%
# 根据zcj降序排序
arrange(desc(zcj)) %>%
# 查看最后两行
tail(n=2)
注意以上操作原数据没有修改, 如果要修改, 使用 %<>% 或者赋值给原变量 <-
选择行
cjb %>%
filter(yw<60) # yw小于60
cjb %>%
# 4~12小于60的行
filter_at(vars(4:12),any_vars(. < 60))
分组统计
cjb %>%
mutate(zcj = rowSums(.[4:12])) %>%
filter(zcj !=0) %>%
group_by(xb) %>% # 基于xb分组
summarise(count=n(),
max = max(zcj),
mean = mean(zcj),
min = min(zcj))
# A tibble: 2 x 5
# xb count max mean min
# <chr> <int> <dbl> <dbl> <dbl>
# 1 女 406 879 797. 647
# 2 男 368 885 793. 523
长宽变换
由于分组变量是列变量
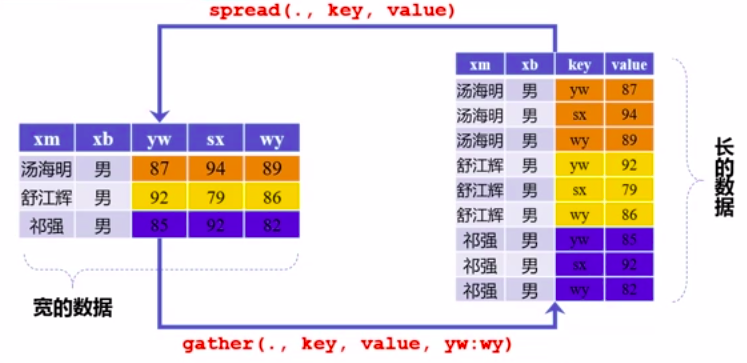
图片alt
cjb %>%
mutate(zcj = rowSums(.[4:12])) %>%
filter(zcj !=0) %>%
group_by(xb) %>% # 基于xb分组
summarise(count=n(),
max = max(zcj),
mean = mean(zcj),
min = min(zcj))
# A tibble: 2 x 5
# xb count max mean min
# <chr> <int> <dbl> <dbl> <dbl>
# 1 女 406 879 797. 647
# 2 男 368 885 793. 523
cjb %>%
mutate(zcj = rowSums(.[4:12])) %>%
filter(zcj!=0) %>%
gather(key = ke_mu,value = cheng_ji,yw:sw) %>%
group_by(ke_mu) %>%
summarise(max = max(cheng_ji),
mean = mean(cheng_ji),
median =median(cheng_ji),
min = min(cheng_ji)) %>%
arrange(desc(mean))
# A tibble: 9 x 5
# ke_mu max mean median min
# <chr> <int> <dbl> <dbl> <int>
# 1 dl 100 93.0 94 70
# 2 zz 100 92.3 93 65
# 3 hx 100 91.7 94 52
# 4 ls 100 89.1 90 0
# 5 wy 99 87.5 88 30
# 6 yw 96 87.4 88 33
# 7 sw 100 86.4 88 55
# 8 sx 100 86.2 89 26
# 9 wl 100 81.2 83 21
x <- formula(". ~ Species")
aggregate(x, data = iris, mean)
subset(iris,Species=="setosa",select="Petal.Width")[[1]] |>mean()
删除重复行
# 根据所有列删除重复的行(完全一样的观测值):
my_data %>% distinct()
# 根据特定列删除重复值
my_data %>% distinct(Sepal.Length, .keep_all = TRUE)
# 根据多列删除重复值
my_data %>% distinct(Sepal.Length, Petal.Width, .keep_all = TRUE)
# 选项.kep_all用于保留数据中的所有变量
注: 本文是学堂在线的笔记
library(tidyverse)
fun <- function(x,y){
cat(x,"---")
str_remove_all(x,y)
}
read_csv("data/GSE168152_hAFs_EV_miRNA_Profile_UMIcounts.csv.gz")[1:5,1:5] |>
column_to_rownames("miRNA") |>
mutate_all(fun)
read_csv("data/GSE168152_hAFs_EV_miRNA_Profile_UMIcounts.csv.gz")[1:5,1:5] |>
column_to_rownames("miRNA") |>
mutate_all(~fun(.,"\'"))
separate_rows
targetScan_all <- Predicted_Targets_Info |>
filter(SpeciesID==9606) |>
separate_rows(miRFamily,sep="/") |>
mutate(miRNA_symbol=case_when(
grepl("miR",miRFamily)~paste0("hsa-",miRFamily),
T~paste0("hsa-miR-",miRFamily)
)) |>
dplyr::select(GeneSymbol,miRNA_symbol) |>
distinct() |>
R语言去除特定列中含有NA的行
library(tidyr)
# 去除表格DE_GENE 中 colnames为gene_id 的列中含有NA的行
DE_GENE <- DE_GENE %>% drop_na(gene_id)
str_split
using strsplit and subset in dplyr and mutate
metadata <- pData(gset)|>
select(geo_accession,title,characteristics_ch1) |>
filter(grepl("microRNA",title)) |>
mutate(group=case_when(grepl("normal",characteristics_ch1)~"control",
grepl("tumor",characteristics_ch1)~"treatment"),
sample = stringr::str_split(title,"_") %>% map_chr(., 1))
https://stackoverflow.com/questions/42565539/using-strsplit-and-subset-in-dplyr-and-mutate
R语言根据一列分组选择另一列最大的行
# 创建示例数据
data <- data.frame(
group = c("A", "A", "B", "B", "C", "C"),
value = c(10, 15, 8, 12, 5, 9)
)
# 根据group列分组,选择value列中最大的行
data %>%
group_by(group) %>%
top_n(1, value)
dplyr
- github
图片alt
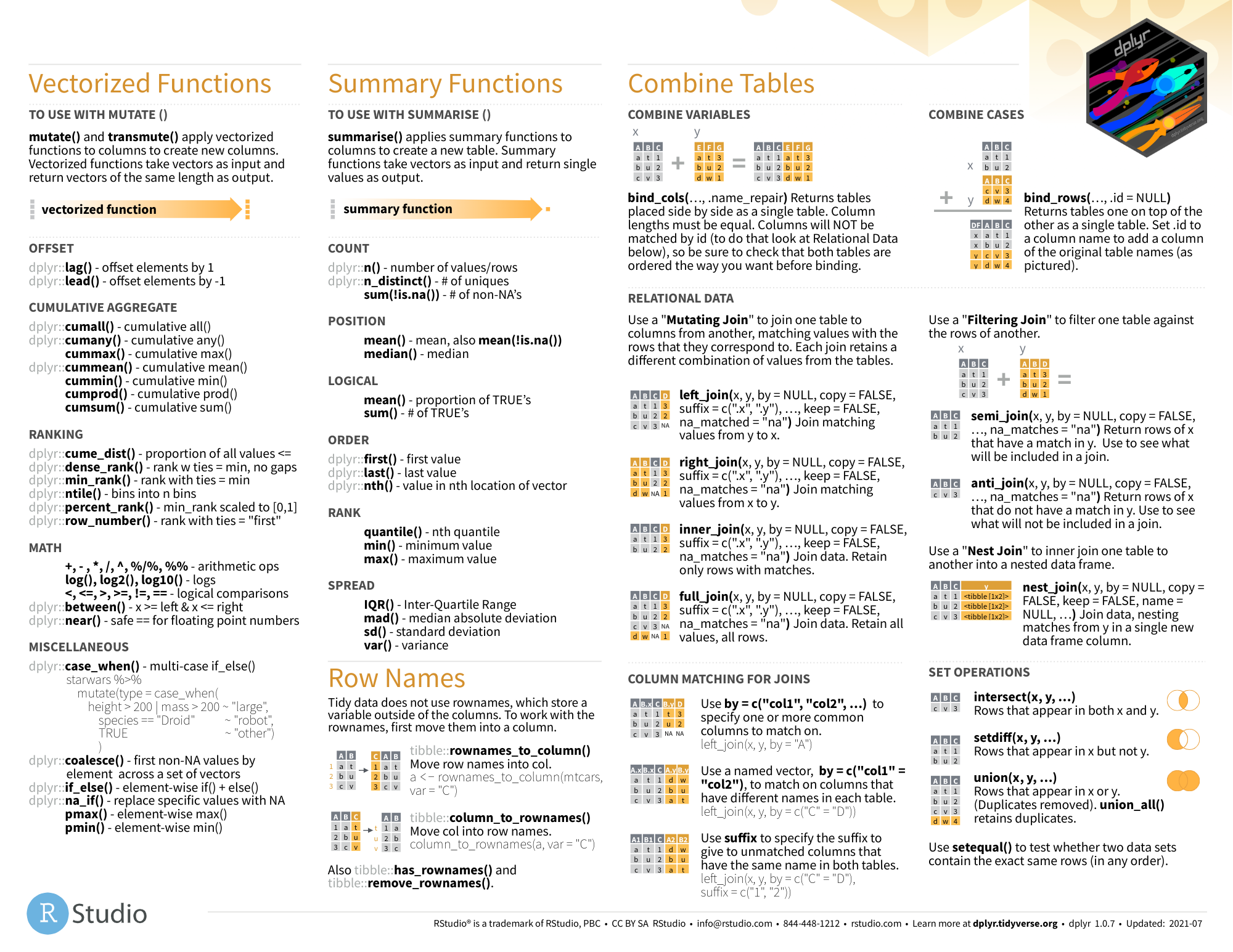
图片alt
tidyr
- github
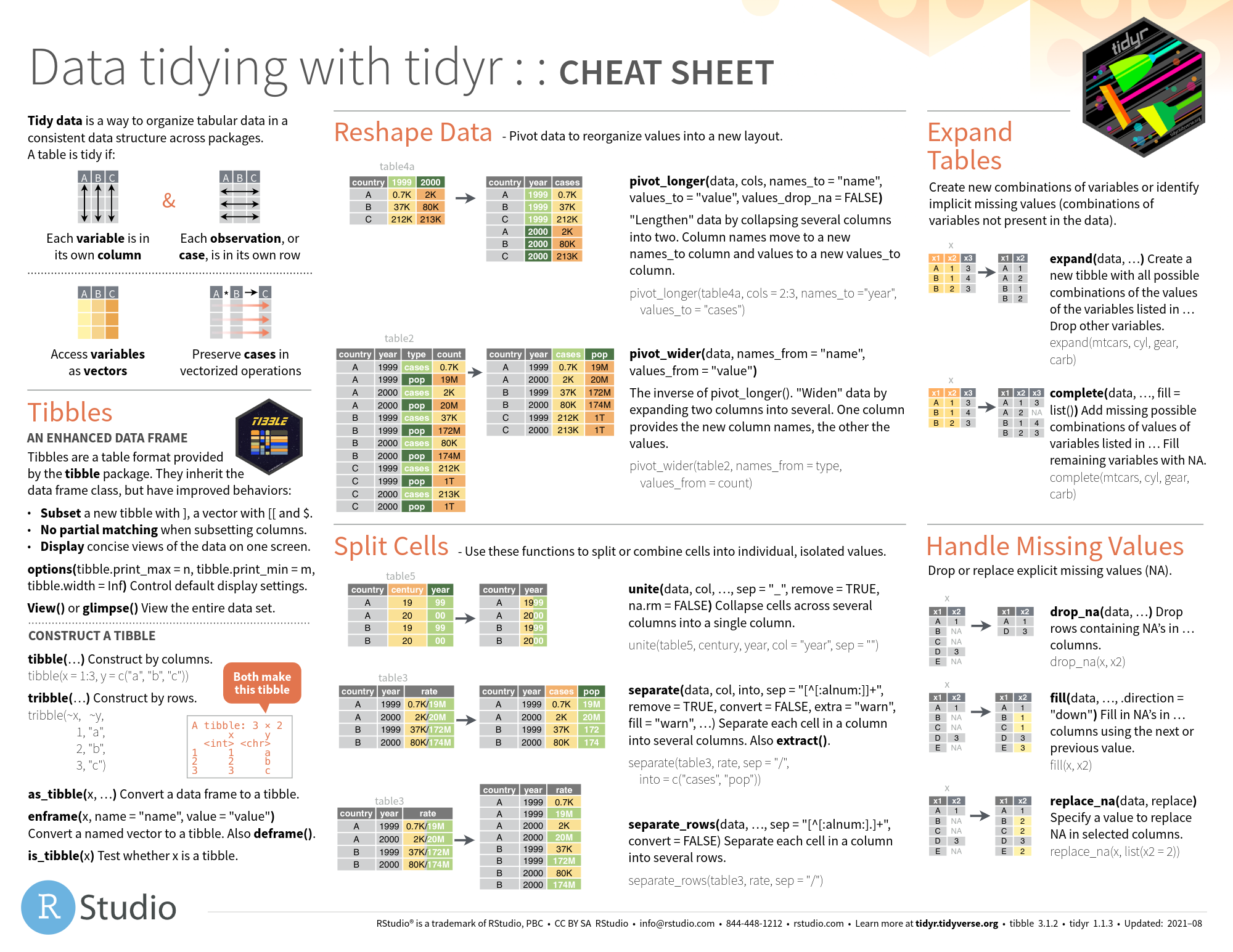
图片alt

图片alt
ggplot2

图片alt
- github

图片alt
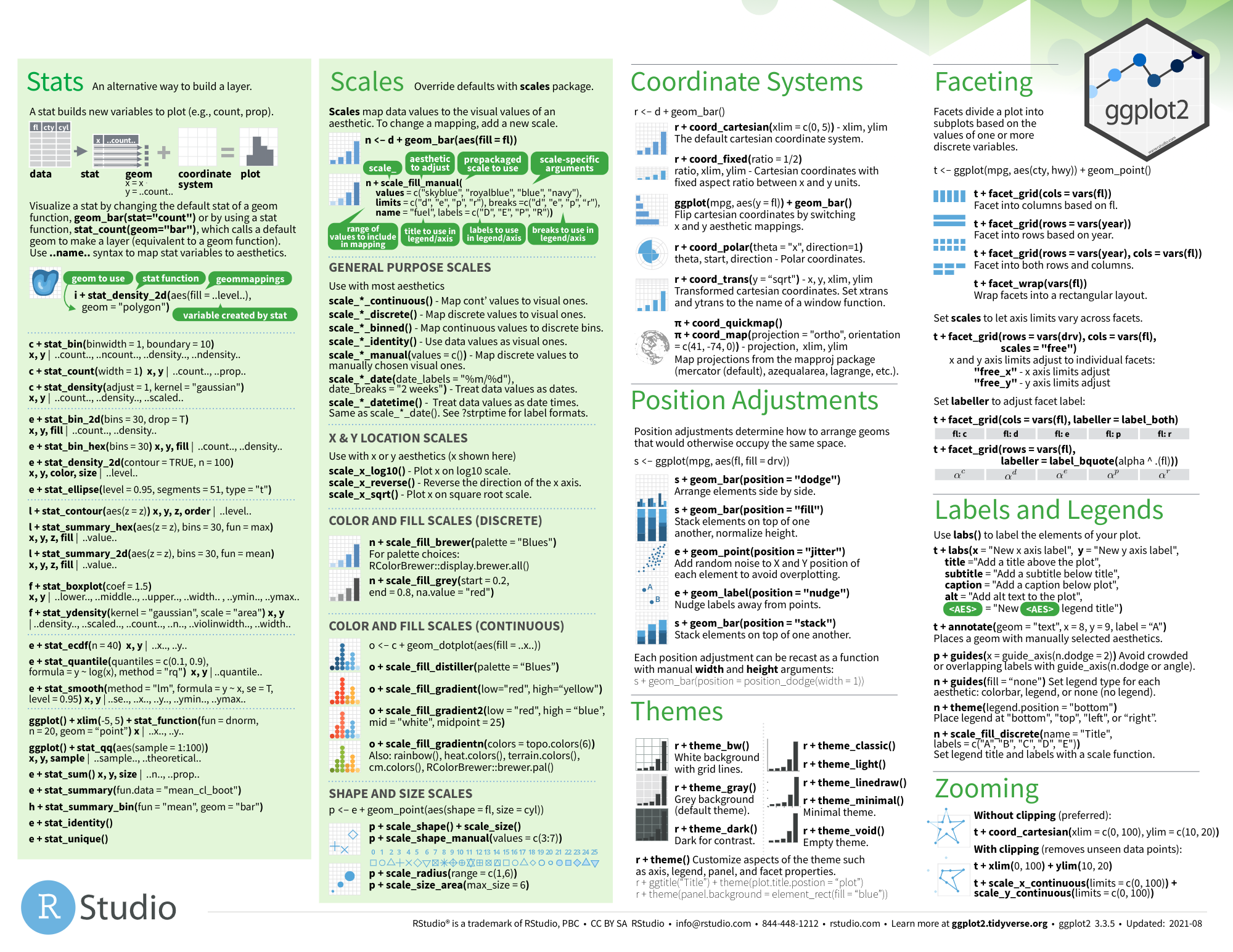
图片alt
purrr
- github

图片alt

图片alt
stringr
- github
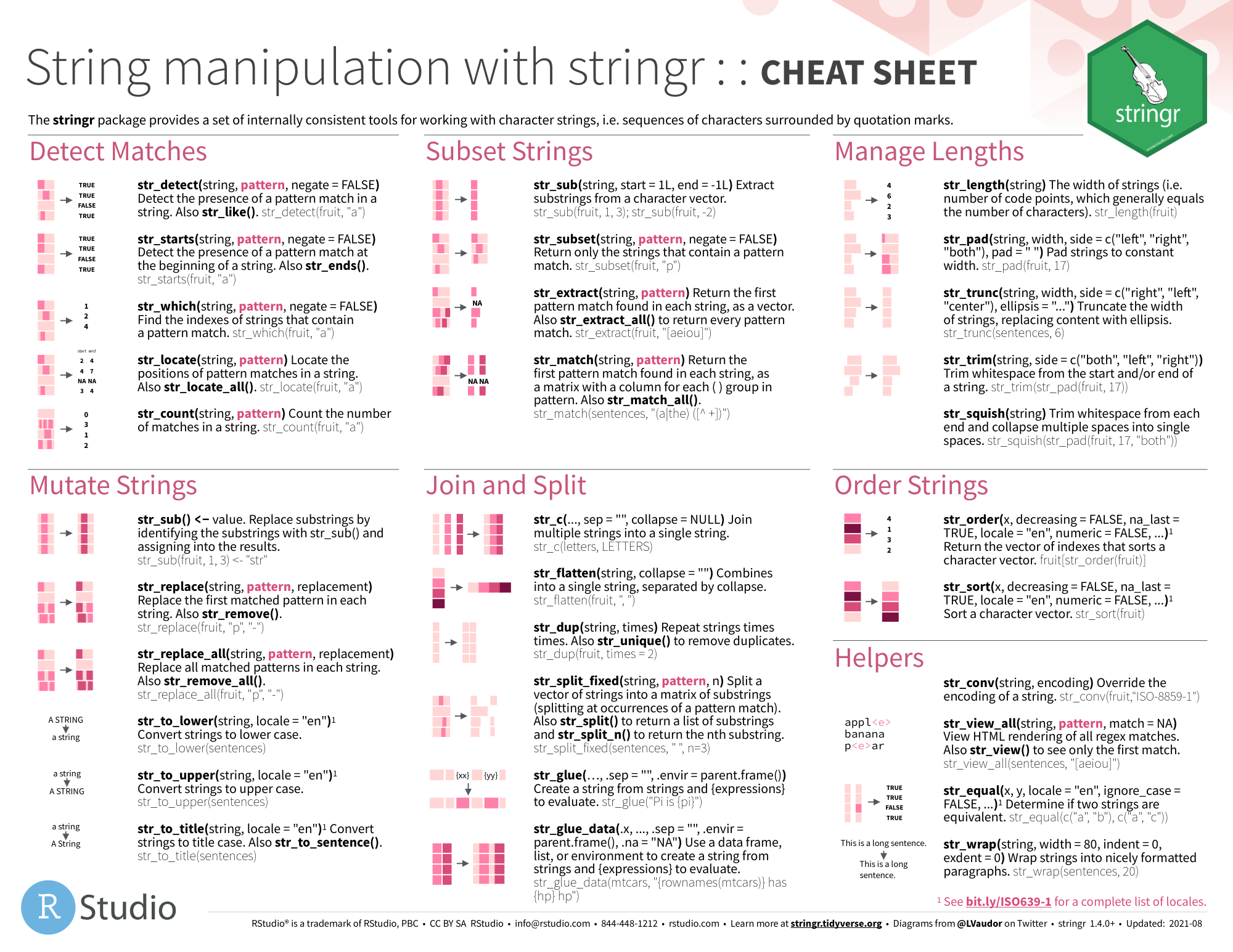
图片alt

图片alt
readr
- github
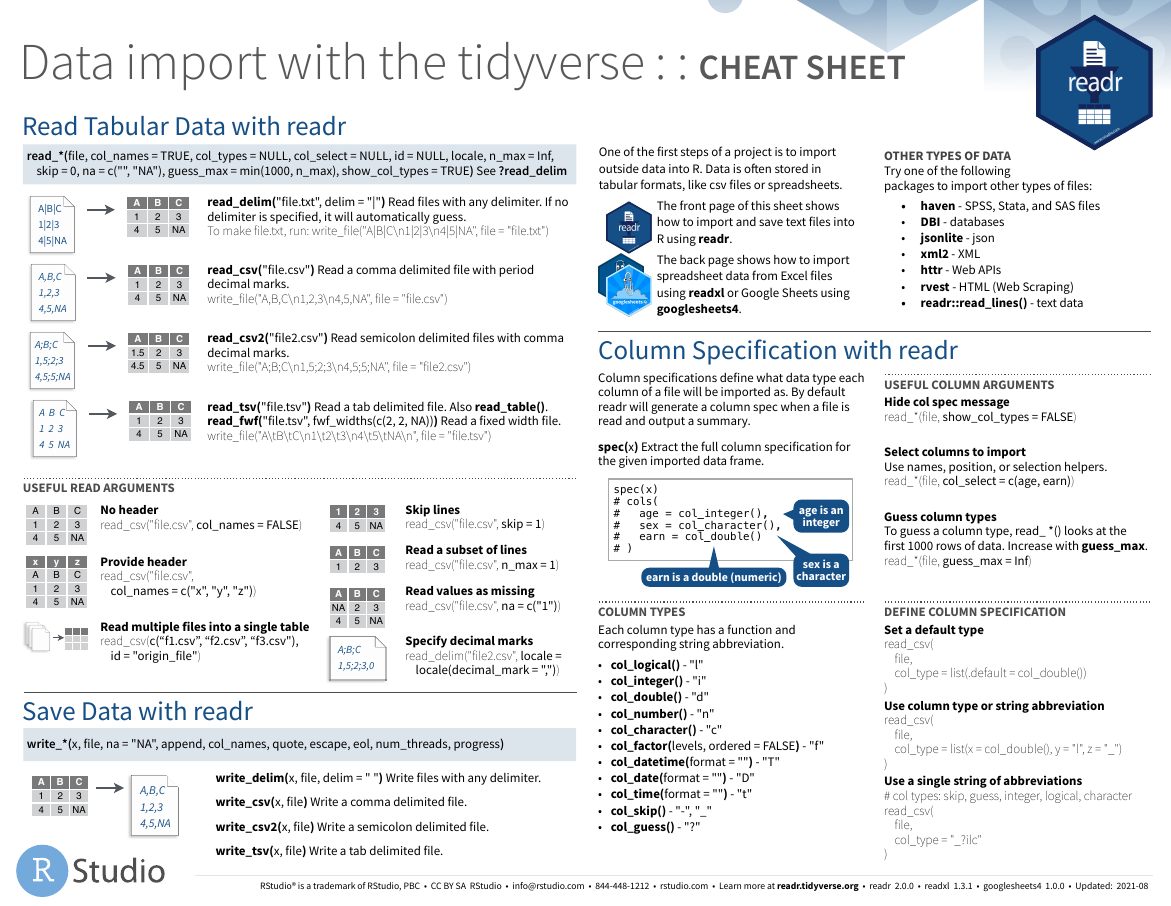
图片alt

图片alt
forcats
- github
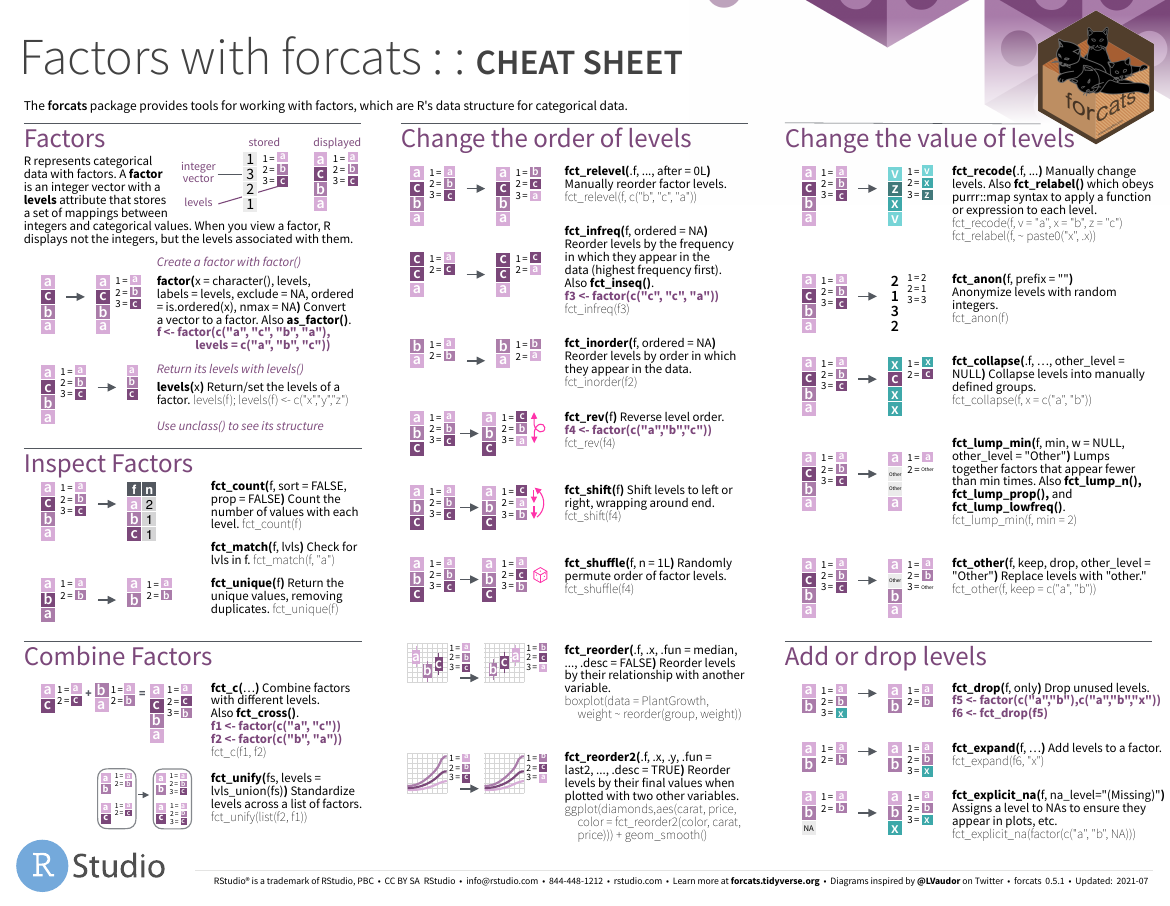
图片alt