- 固定参数函数
- 可变参数的函数
- 一切都是对象
- 泛型和多态
- R递归
- 匿名函数
- 循环
- tidyverse重命名列名(rename)
- 通过正则表达式截取字符串
- tidyverse
- R语言移除data frame所有空格
- 计算行的个数
- R语言NA处理
- 重命名列
- Reduce
- 数据统计
- assign function
- tidyverse用字符串传递变量名称和列名
- Map 和 Reduce
- 参考
固定参数函数
f1 <- function(a,b=2){
message(a,b)
return(a+b) # 没有return返回最后一条语句
}
b <- f1(5) # 52
b # 7
f1(2,5) # 25
f1(b=2,a=5) # 52
f1(b=5,2) # 25
可变参数的函数
r自带函数 c(..., recursive = FALSE, use.names = TRUE)
f2 <- function(...){
cat(..2) # 2
dot_args = list(...)
print(dot_args)
}
f2(1,2,3,4,5)
一切都是对象
操作符也是函数类型对象
1+2 # 3
"+"(1,2) # 3
`+`(1,2) # 3
'+'(1+2) # 3
'<-'(new_var,5)
new_var # 5
':'(1,5) # 1 2 3 4 5
'['(1:10,2) # 2
自定义运算符
注意:字母作为名称的函数必须使用%%括住名称,才能放在中间被调用
'%ab2c%' <- function(a,b){
sqrt(sum(a^2,b^2))
}
ab2c(3,4) # Error in ab2c(3, 4) : could not find function "ab2c"
3 %ab2c% 4
纯粹的字母不能在两个参数之间当做函数使用
'ab2c' <- function(a,b){
sqrt(sum(a^2,b^2))
}
ab2c(3,4) # [1] 5
3 %ab2c% 4 # Error in 3 %ab2c% 4 : could not find function "%ab2c%"
ab2c <- function(a,b){
sqrt(sum(a^2,b^2))
}
ab2c(3,4) # [1] 5
3 %ab2c% 4 # Error in 3 %ab2c% 4 : could not find function "%ab2c%"
函数名称是符号可以不使用%%, 但是使用%%和不使用%%是不同的函数
5+2 # 7
"+" <- function(x,y){
x*y
}
"%+%" <- function(x,y){
x/y
}
5+2 # 10
5 %+% 2 # [1] 2.5
rm(`+`)
5+2 # 7
对于%>%的理解
library(purrr)
x <- c(1,1,2,2,8,5,9)
x %>%
unique() %>%
sort()
# > 1 2 8 5 9
x <- unique(x)
x <- sort(x)
# > 1 2 8 5 9
泛型和多态
函数接受不同的对象有不同的结果
x <- seq(1,100,by=10)
y <- 2*x + 10
xy <- cbind(x,y)
class(xy) # matrix
plot(xy,
xlim = c(1,100),
ylim = c(0,230),
type = "o",col="red") # figure 1
x <- seq(1,100,by=10)
y <- 2*x + 10
xy <- cbind(x,y)
my_model <- lm(y~x)
class(my_model) # lm
op <- par(mfrow = c(2,2))
plot(my_model) # figure 2
par(op)
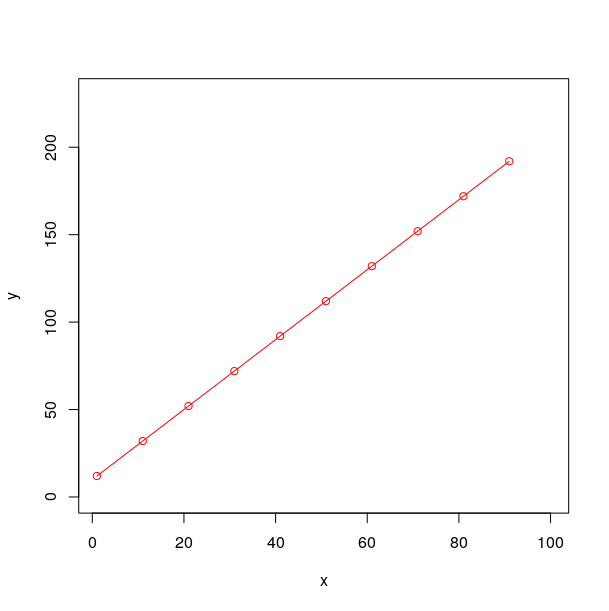 figure 1
figure 1
 figure 2
figure 2
自定义泛型函数
my_function <- function(x,y){
message("I am interface")
UseMethod("my_function",x) # 通过x标签类型确定执行函数
}
my_function.addXY <- function(x,y){
return(x+y)
}
my_function.multiplyXY <- function(x,y){
return(x*y)
}
my_function.default <- function(x,y){
return(x-y)
}
x<-9
y<-5
my_function(x,y) # default 4
class(x) <- "addXY"
my_function(x,y) # addXY 14
class(x) <- "multiplyXY"
my_function(x,y) # multiplyXY 45
对于泛型函数"+"的理解
"+" 是有两个参数函数的操作
"+.onlyFirst" <- function(a,b){
return(a[1]+b[1])
}
a <- 1:5
b <- 6:10
a+b # 7 9 11 13 15
"+"(a,b) # 7 9 11 13 15
class(a) <- "onlyFirst"
a+b # 7
"+"(a,b) # 7
class(a) <- NULL
class(b) <- "onlyFirst"
a+b # 7
"+"(a,b) # 7
class(a) <- "onlyFirst"
class(b) <- "onlyFirst"
a+b # 7
"+"(a,b) # 7
R递归
fun1 <- function(s){
message("before",s)
if(s>0){
fun1(s-1)
}
message("after",s)
}
fun1(5)
before5
before4
before3
before2
before1
before0
after0
after1
after2
after3
after4
after5
递归Fibonacci
fib <- function(n){
if(n==1){
return(1)
}else{
return(c(fib(n-1),sum(fib(n-1),n=2)))
}
}
fib(10)
# > 1 3 6 12 24 48 96 192 384 768
匿名函数
split_df <- function(starbase_miRNA_data){
tmp<-strsplit(starbase_miRNA_data$miRNA,split="-",fixed=TRUE)
starbase_miRNA_data <- sapply(tmp,function(x) paste(x[1:3],collapse = "-"))
data.frame(miRNA=unique(starbase_miRNA_data))
}
gset_1_pd <- pData(gset_1) %>%
dplyr::select(sample_id=geo_accession,sample_name=title,tissue_type=source_name_ch1)%>%
mutate(group=str_sub(tissue_type,-6,-1))
gset_1_pd <- pData(gset_1) %>%
dplyr::select(sample_id=geo_accession,sample_name=title,tissue_type=source_name_ch1)%>%
mutate(group= change_col(tissue_type)
change_col <- function(x){
return(paste0("a",x))
}
gset_1_pd <- pData(gset_1) %>%
dplyr::select(sample_id=geo_accession,sample_name=title,tissue_type=source_name_ch1)%>%
mutate(group= tissue_type)%>%
mutate_at(vars("group"),~ change_col(.))
change_col <- function(x){
return(paste0("a",x))
}
循环
apply(a,1, function(x) mean(x))
rowMeans(a``)
tidyverse重命名列名(rename)
plyr::rename(data, c(old=new))
dplyr::rename(data, new = old)
通过正则表达式截取字符串
str_extract(miRNA,"hsa-let-[0-9]+[a-z]?")
tidyverse
- add_count
- mutate(type = str_c(direction,type,sep="_"))
R语言移除data frame所有空格
as.data.frame(apply(mutation_maf,2,function(x){trimws(x, which = c("both"))}))
df1$Remove_all_space <- gsub('\\s+', '', df1$State)
https://stackoverflow.com/questions/20760547/removing-whitespace-from-a-whole-data-frame-in-r
计算行的个数
# library(data.table)
# data.table(mutation_maf)->mutation_maf_tab
# mutation_maf_tab[,.N, .(Hugo_Symbol, Variant_Classification)] %>%
# filter(Hugo_Symbol=="TP53")
R语言NA处理
idx <- which(!complete.cases(tcga_clinical))
tcga_clinical[idx,]
subset(tcga_clinical,!complete.cases(tcga_clinical))
na.omit(tcga_clinical)%>%dim()
tcga_clinical%>%dim()
重命名列
iris %>%
set_names("sep_len", "sep_wid", "pet_len", "pet_wid", "spp") %>%
names()
Reduce
circRNA P.Value logFC
1 hsa_circRNA_101175 0.0000825 -1.17
2 hsa_circRNA_002178 0.00011 1.91
3 hsa_circRNA_000554 0.000132 -1.62
4 hsa_circRNA_101004 0.000141 -1.33
5 hsa_circRNA_001678 0.000163 2.68
6 hsa_circRNA_101748 0.000277 1.28
intersect_circRNA <- Reduce(function(x,y) merge(x,y,by="circRNA",all=F),list(gse_1,gse_2,gse_3),accumulate=F)
图片alt
数据统计
m <- data.frame(matrix(sample(100, 20, replace = TRUE), ncol = 4))
m
colMeans(m)
rowMeans(m)
colSums(m)
summarise(m,count=n(),max = max(m), mean = mean(m))
summary(m)
Reduce(function(x,y) intersect(x,y),list(circbank_miRNA,miRanda_miRNA,RNAhybrid_mimRNA),accumulate =FALSE)
将b的顺序调整为a
a<- c("a","b","c")
b <-c("b","c","a")
b[match(a,b)]
data.matrix <- do.call('cbind',lapply(gsmlist,function(x)
{tab <- Table(x)
mymatch <- match(probesets,tab$ID_REF)
return(tab$VALUE[mymatch])
}))
https://bioconductor.org/packages/release/bioc/vignettes/GEOquery/inst/doc/GEOquery.html
suppressMessages(library(tidyverse))
getName <- function(x,res){
last <- x[length(x)]
if(!grepl("Unclassified",last, ignore.case=T)){
res <- c(res,last)
}
if(length(res)>1 || length(x) ==1){
return(res)
}
getName(x[1:length(x)-1],res)
}
lapply(list.files("/data/metagenomics/mtbi_out/micop_virus/",pattern = "merged_abundance_table_(.*)\\D.txt"),function(x){
read_tsv(x,show_col_types = FALSE,comment="#") |>
mutate(TAXPATHSN=str_split(TAXPATHSN,"\\|") %>% map_chr(., function(x){
name <- paste0(getName(x,c()),collapse = "_")
return(name)
})) %>% {.[!duplicated(.$TAXPATHSN),]} |> head() #|> write_tsv(file = "")
})
assign function
可以动态创建变量
for(i in 1:3) { # Head of for-loop
assign(paste0("x_", i), i) # Combining assign & paste0
}
assign("goAnno", goAnno, envir=GO_Env)
assign("keytype", keytype, envir=GO_Env)
assign("ont", ont, envir = GO_Env)
assign("organism", get_organism(OrgDb), envir=GO_Env)
tidyverse用字符串传递变量名称和列名
如:如果n是一个变量名称,后又定义x <- "n",而之后的变量n的引用想要使用x来表述,这时候x代表的是字符串"n",而不是变量n。
当定义的字符串向量要传递给tidyverse 时,字符传递给tidyverse 时可能会有问题。因为tidyverse 中列名通常是不带引号的。
解决办法:使用get()或者!! 方法;如果是ggplot2中的映射,可以使用aes_string()。
df %>% select(!!x) %>% head()
## Sepal.Length
## 1 5.1
## 2 4.9
## 3 4.7
## 4 4.6
## 5 5.0
## 6 5.4
https://zhuanlan.zhihu.com/p/559652326
Map 和 Reduce
Reduce(function(x,y)intersect(x,y),cnames)
Map(identical,list( cnames[[1]]),cnames)
参考
https://blog.csdn.net/weixin_29534143/article/details/113581866