- 数据准备
- 使用训练集建立模型,并查看训练集的混淆矩阵和ROC曲线
- 使用训练集建立模型,并查看测试集的混淆矩阵和ROC曲线
- 使用交叉验证建立随机森林的二分类模型
- Mean Decrease Accuracy and Mean Decrease Gini
- 关于ROC曲线上的每一个点
- 参考
在随机森林方法中,创建了大量决策树。每个观察结果都被输入到每个决策树中。每个观察的最常见结果用作最终输出。新的观察结果被输入到所有树中,并对每个分类模型进行多数表决。
数据准备
加载R包
library(randomForest)
这里使用R语言的自带数据集iris,选择versicolor和virginica两种类型用于二分类问题
data(iris)
iris <- dplyr::filter(iris,Species %in% c("versicolor","virginica"))
iris$Species<-as.factor(as.character(iris$Species))
head(iris)
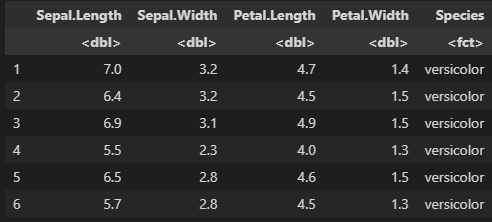
图片alt
使用训练集建立模型,并查看训练集的混淆矩阵和ROC曲线
set.seed(2022)
iris.rf<-randomForest(Species ~ ., data=iris,ntree=10,importance=T,proximity=T)
print(iris.rf)
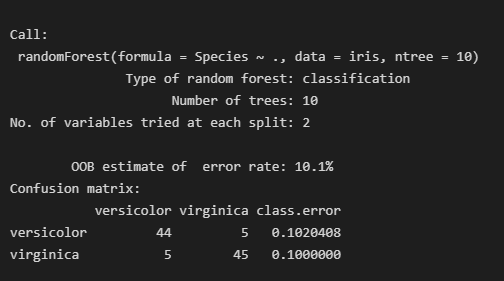
图片alt
查看混淆矩阵
data.frame(predicted =iris.rf$predicted,
group = iris$Species,
stringsAsFactors = F)|> table()
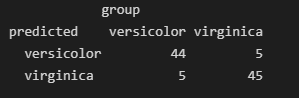
图片alt
data.frame(predicted =predict(iris.rf, newdata = iris,type="response"),
group = iris$Species,
stringsAsFactors = F)|> table()
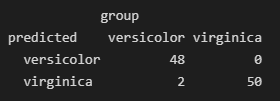
图片alt
绘制ROC曲线只需要简单的下面一行代码
library(pROC)
plot.roc(iris$Species,iris.rf$votes[,2])
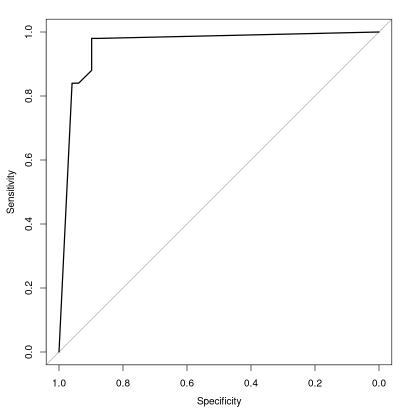
图片alt
下面的ROC曲线添加了图例和颜色
library(RColorBrewer)
mycol<-brewer.pal(11, "Spectral")
x <- plot.roc(iris$Species,iris.rf$votes[,2],ylim=c(0,1),xlim=c(1,0),
smooth=FALSE,
ci=TRUE,
col=mycol[1],
lwd=2,
legacy.axes=T)
ci.lower <- round(as.numeric(x$ci[1]),3)
ci.upper <- round(as.numeric(x$ci[3]),3)
legend.name <- paste("AUC:",round(as.numeric(x$auc),3),paste(ci.lower,ci.upper,sep="-"))
legend("bottomright",
legend=legend.name,
col = mycol[1],
lwd = 2,
bty="n")
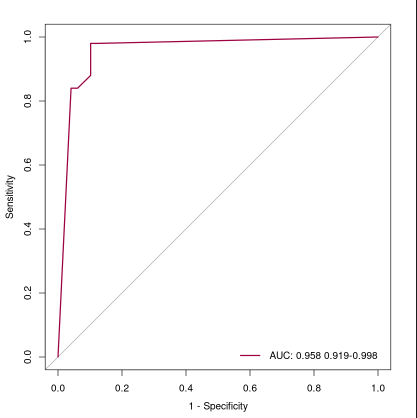
图片alt
使用训练集建立模型,并查看测试集的混淆矩阵和ROC曲线
将数据集划分成70%的训练集和30%的测试集
library(caTools)
set.seed(2022)
sample <- sample.split(iris$Species, SplitRatio = 0.7)
train <- subset(iris, sample == TRUE)
test <- subset(iris, sample == FALSE)
建立二分类器的随机扥林模型
iris.rf<-randomForest(Species ~ ., data=train,ntree=10)
print(iris.rf)
iris.rf$importance

图片alt
查看混淆矩阵
data.frame(predicted =predict(iris.rf, newdata = test,type="response"),
group = test$Species,
stringsAsFactors = F)|> table()
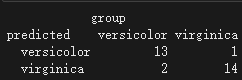
图片alt
绘制ROC曲线
library(pROC)
plot.roc(test$Species,predict(iris.rf, newdata = test,type="vote")[,2])
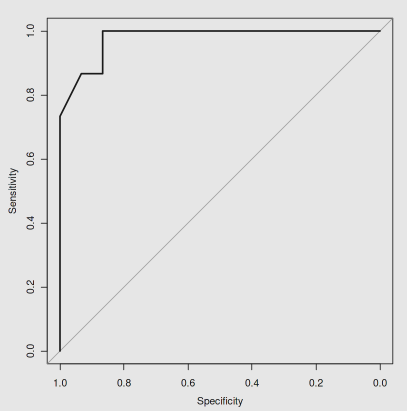
图片alt
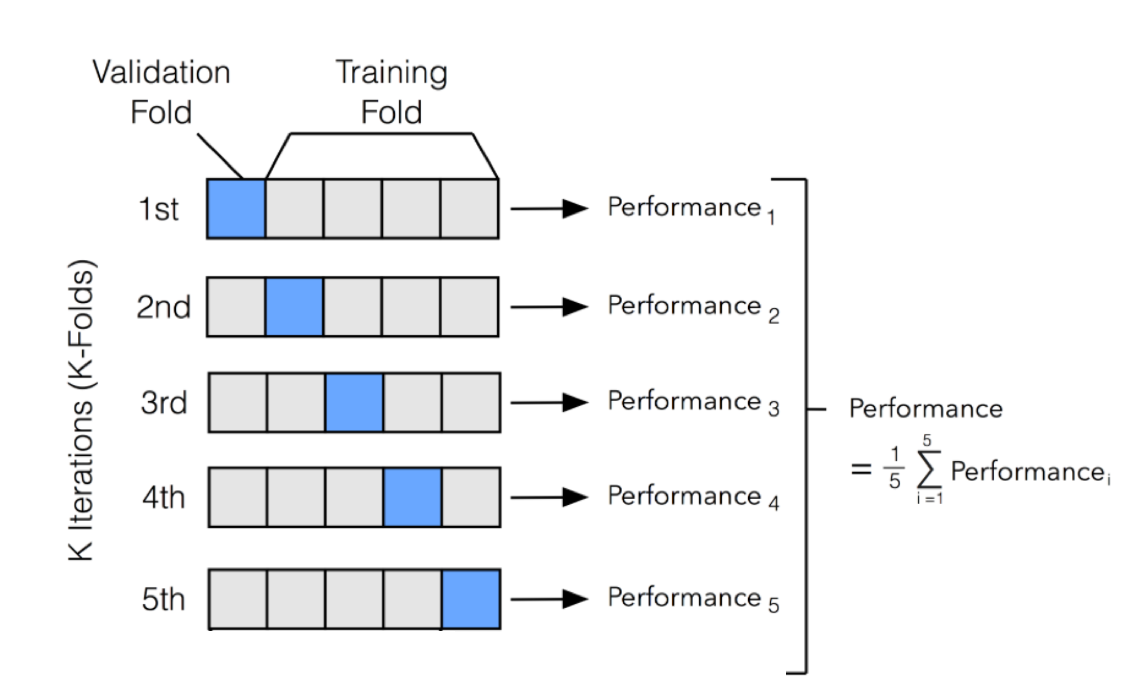
使用交叉验证建立随机森林的二分类模型
将数据集划分为大小相同的k份
每一次将其中一份作为测试集,剩余的k-1份作为训练集
以k次测试结果的平均值作为最终的测试误差
cv_kfold <- function (data,k=10,seed=2022){
n_row <- nrow(data)
n_foldmarkers <- rep(1:k, ceiling(n_row/k))[1:n_row]
set.seed(seed)
n_foldmarkers <- sample(n_foldmarkers)
k_fold <- lapply(1:k,function (i){
(1:n_row)[n_foldmarkers==i]
})
return(k_fold)
}
图片alt
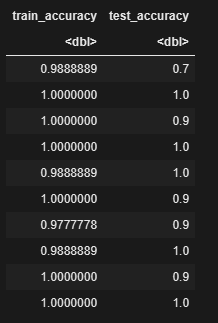
res <- data.frame()
for (i in 1:length(kfolds)){
curr_fold <- kfolds[[i]]
train_set <- iris[-curr_fold,]
test_set <- iris[curr_fold,]
predicted_train <-randomForest(Species ~ ., data=train_set,ntree=10)
train_accuracy <- sum(train_set$Species ==predict(predicted_train, newdata = train_set,type="response")) / length(train_set$Species)
test_accuracy <- sum(test_set$Species ==predict(predicted_train, newdata = test_set,type="response") )/length(test_set$Species)
res <- rbind(res,data.frame(train_accuracy=train_accuracy, test_accuracy=test_accuracy))
}
res
图片alt
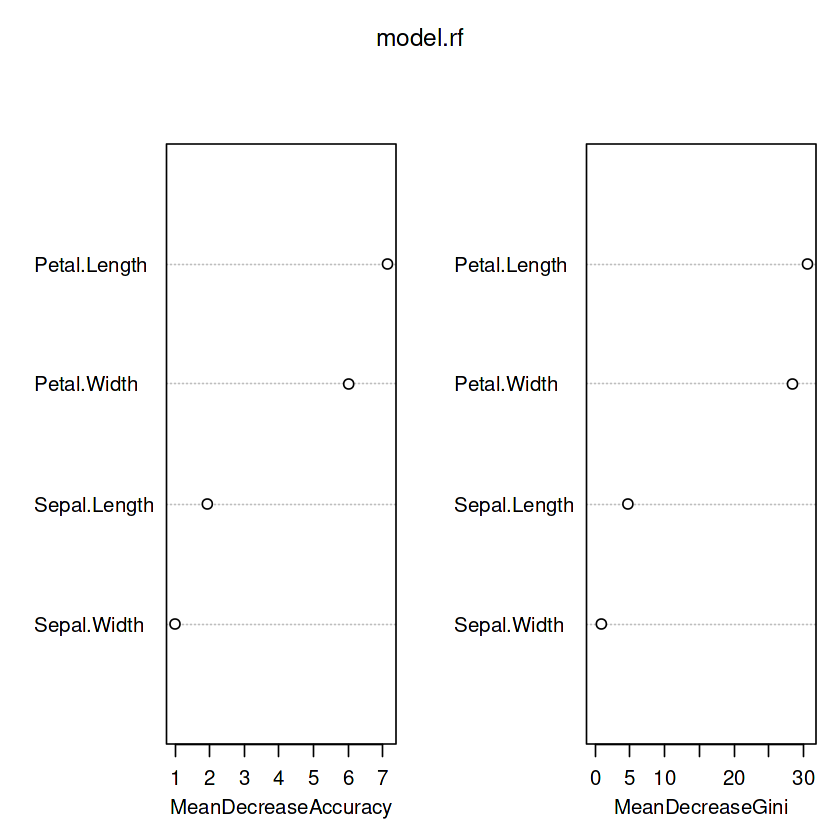
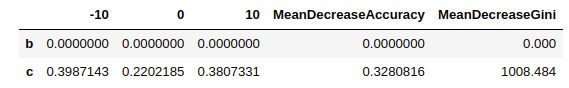
Mean Decrease Accuracy and Mean Decrease Gini
https://stackoverflow.com/questions/55036416/why-is-mean-decrease-gini-in-random-forest-dependent-on-population-size/55055245#55055245
varImpPlot(model.rf)
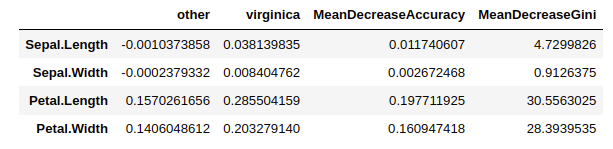
model.rf$importance
set.seed(1)
n <- 1000
# There are three classes in equal proportions
a <- rep(c(-10,0,10), each = n)
# One feature is useless
b <- rnorm(3*n)
# The other feature is highly predictive but we need at least two splits
c <- rnorm(3*n, a)
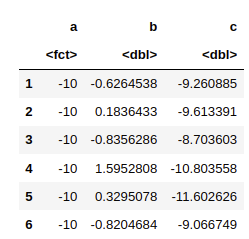
data <- data.frame(a = as.factor(a), b = b, c = c)
head(data)
rf3 <- randomForest(data = data, a ~ b + c, importance = TRUE,
ntree = 1000, mtry = 2, maxnodes = 2)
rf3$importance
rf3 <- randomForest(data = data, a ~ b + c, importance = TRUE,
ntree = 1000, mtry = 2, maxnodes = 3)
rf3$importance

关于ROC曲线上的每一个点
library(caTools)
set.seed(2022)
sample <- sample.split(iris$Species, SplitRatio = 0.9)
train <- subset(iris, sample == TRUE)
test <- subset(iris, sample == FALSE)
iris.rf<-randomForest(Species ~ ., data=train,ntree=10)
prob <- predict(iris.rf, newdata = test,type="vote")
predicted <- predict(iris.rf, newdata = test,type="response")
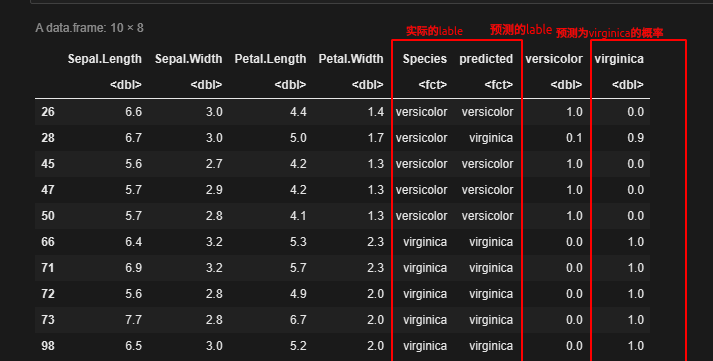
cbind(test,predicted, prob)
图片alt
绘制ROC曲线
plot.roc(test$Species,prob[,2])
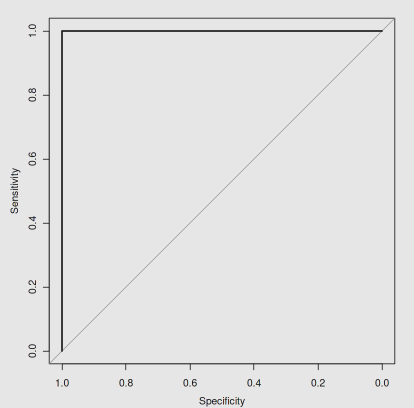
ROC曲线上的每一个点表示不同阈值下
- 模型预测的假阳性率或称为敏感性(sensitivity, False Positive Rate, FPR),即对应图中的横坐标,该值越小,模型的性能越好
- 模型预测的特异性或称为真阳性率(specificity, True Positive Rate, TPR),即对应图中的纵坐标,该值越大,模型性能越好
假阳性率=预测结果中负样本个数 / 负样本的个数
真阳性率=预测结果中正样本个数 / 正样本个数
这里的阈值其实就是prob[,2],即预测为virginca的概率
当阈值取0时,也就是说预测概率大于等于0时,我们将样本预测为virginca,上表中大于零的样本全部预测正确,即纵坐标真阳性率的值为1;小于等于0的全部预测为versicolor,即假阳性率为0,此时得到一个点(0,1)
然后依次按照上表virginica这一列为阈值,计算不同阈值下的假阳性率和真阳性率,得到10个点,再将10个点连起来,就构成ROC曲线