Molecular Signatures Database is a collection of annotated gene sets. It contains 8 major collections:
- H: hallmark gene sets
- C1: positional gene sets
- C2: curated gene sets
- C3: motif gene sets
- C4: computational gene sets
- C5: GO gene sets
- C6: oncogenic signatures
- C7: immunologic signatures
Users can download GMT files from Broad Institute and use the read.gmt() function to parse the file to be used in enricher() and GSEA().
There is an R package, msigdbr, that already packed the MSigDB gene sets in tidy data format that can be used directly with clusterProfiler (Yu et al. 2012).
library(msigdbr)
msigdbr_show_species()
## [1] "Anolis carolinensis" "Bos taurus"
## [3] "Caenorhabditis elegans" "Canis lupus familiaris"
## [5] "Danio rerio" "Drosophila melanogaster"
## [7] "Equus caballus" "Felis catus"
## [9] "Gallus gallus" "Homo sapiens"
## [11] "Macaca mulatta" "Monodelphis domestica"
## [13] "Mus musculus" "Ornithorhynchus anatinus"
## [15] "Pan troglodytes" "Rattus norvegicus"
## [17] "Saccharomyces cerevisiae" "Schizosaccharomyces pombe 972h-"
## [19] "Sus scrofa" "Xenopus tropicalis"
We can retrieve all human gene sets:
m_df <- msigdbr(species = "Homo sapiens")
head(m_df, 2) %>% as.data.frame
## gs_cat gs_subcat gs_name gene_symbol entrez_gene ensembl_gene
## 1 C3 MIR:MIR_Legacy AAACCAC_MIR140 ABCC4 10257 ENSG00000125257
## 2 C3 MIR:MIR_Legacy AAACCAC_MIR140 ABRAXAS2 23172 ENSG00000165660
## human_gene_symbol human_entrez_gene human_ensembl_gene gs_id gs_pmid
## 1 ABCC4 10257 ENSG00000125257 M12609
## 2 ABRAXAS2 23172 ENSG00000165660 M12609
## gs_geoid gs_exact_source gs_url
## 1
## 2
## gs_description
## 1 Genes having at least one occurence of the motif AAACCAC in their 3' untranslated region. The motif represents putative target (that is, seed match) of human mature miRNA hsa-miR-140 (v7.1 miRBase).
## 2 Genes having at least one occurence of the motif AAACCAC in their 3' untranslated region. The motif represents putative target (that is, seed match) of human mature miRNA hsa-miR-140 (v7.1 miRBase).
Or specific collection. Here we use C6, oncogenic gene sets as an example:
m_t2g <- msigdbr(species = "Homo sapiens", category = "C6") %>%
dplyr::select(gs_name, entrez_gene)
head(m_t2g)
## # A tibble: 6 × 2
## gs_name entrez_gene
## <chr> <int>
## 1 AKT_UP_MTOR_DN.V1_DN 25864
## 2 AKT_UP_MTOR_DN.V1_DN 95
## 3 AKT_UP_MTOR_DN.V1_DN 137872
## 4 AKT_UP_MTOR_DN.V1_DN 134
## 5 AKT_UP_MTOR_DN.V1_DN 55326
## 6 AKT_UP_MTOR_DN.V1_DN 55326
MSigDb over-presentaton analysis
em <- enricher(gene, TERM2GENE=m_t2g)
head(em)

图片alt
MSigDb gene set enrichment analysis
In over-presentaton analysis, we use oncogenic gene sets (i.e. C6) to test whether the DE genes are involved in the process that leads to cancer. In this example, we will use the C3 category to test whether genes are up/down-regulated by sharing specific motif using GSEA approach.
C3_t2g <- msigdbr(species = "Homo sapiens", category = "C3") %>%
dplyr::select(gs_name, entrez_gene)
head(C3_t2g)
## # A tibble: 6 × 2
## gs_name entrez_gene
## <chr> <int>
## 1 AAACCAC_MIR140 10257
## 2 AAACCAC_MIR140 23172
## 3 AAACCAC_MIR140 81
## 4 AAACCAC_MIR140 81
## 5 AAACCAC_MIR140 90
## 6 AAACCAC_MIR140 8754
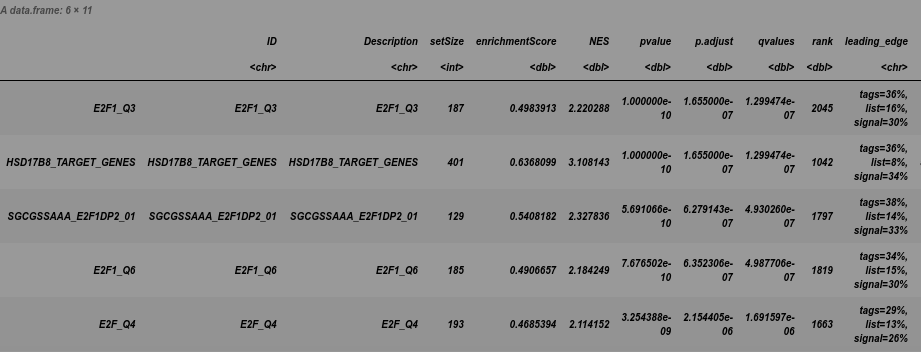
em2 <- GSEA(geneList, TERM2GENE = C3_t2g)
head(em2)
图片alt