Cell Marker
cell_marker_data <- vroom::vroom('http://bio-bigdata.hrbmu.edu.cn/CellMarker/download/Human_cell_markers.txt')
## instead of `cellName`, users can use other features (e.g. `cancerType`)
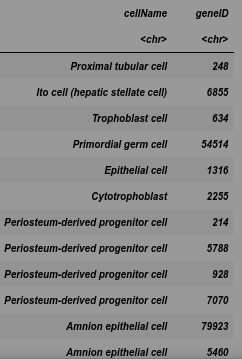
cells <- cell_marker_data %>%
dplyr::select(cellName, geneID) %>%
dplyr::mutate(geneID = strsplit(geneID, ', ')) %>%
tidyr::unnest()
图片alt
Cell Marker over-presentaton analysis
x <- enricher(gene, TERM2GENE = cells)
setReadable(x,OrgDb = "org.Hs.eg.db",keyType = "ENTREZID") |> head()

图片alt
Cell Marker gene set enrichment analysis
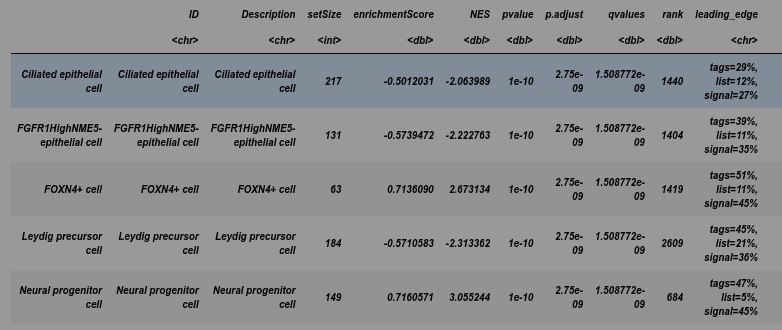
y <- GSEA(geneList, TERM2GENE = cells)
head(y)
图片alt
参考
- CellMarker: a manually curated resource of cell markers in human and mouse
- http://xteam.xbio.top/CellMarker/download.jsp
- http://yikedaxue.slwshop.cn/download.php
- http://yulab-smu.top/biomedical-knowledge-mining-book/universal-api.html