2021.6.27
HTSeq - Counts

图片alt
HTSeq - FPKM

图片alt
BCGSC miRNA Profiling
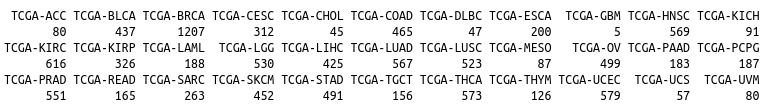
图片alt
相关代码
library(TCGAbiolinks)
project_df <- (function(){
projects <- TCGAbiolinks:::getGDCprojects()$project_id
projects_full <- projects[grepl('^TCGA',projects,perl=T)]
projects_ID <- sapply(projects_full,function(x){stringi::stri_sub(x,6,10)})
res <- data.frame(cancer=projects_ID,TCGA_ID=projects_full)
return(res)
})()
match.file.cases.all <- NULL
for(project in project_df[,2]){
#project <- project_df[5,2]
query<- GDCquery(project = project,
data.category = "Transcriptome Profiling",
data.type = "Gene Expression Quantification",
workflow.type = "HTSeq - Counts")
match.file.cases <- getResults(query,cols=c("cases","file_name"))
match.file.cases$project <- project
match.file.cases.all <- rbind(match.file.cases.all,match.file.cases)
}
readr::write_tsv(match.file.cases.all, path = "filename_case.txt")
table(match.file.cases.all$project)
query<- GDCquery(project = project,
data.category = "Transcriptome Profiling",
data.type = "Gene Expression Quantification",
workflow.type = "HTSeq - FPKM")
query<- GDCquery(project = project,
data.category = "Transcriptome Profiling",
data.type = "miRNA Expression Quantification",
workflow.type = "BCGSC miRNA Profiling")