使用plotly或者dash进行数据可视化
Installation
pip install plotly==4.14.3
import plotly.graph_objects as go
fig = go.Figure(data=go.Bar(y=[2, 3, 1]))
fig.write_html('first_figure.html', auto_open=True)
JupyterLab Support
pip install jupyterlab "ipywidgets>=7.5"
import plotly.graph_objects as go
fig = go.Figure(data=go.Bar(y=[2, 3, 1]))
fig.show()
图片alt
Static Image Export
pip install -U kaleido
usage dash
import plotly.graph_objects as go # or plotly.express as px
fig = go.Figure(data=go.Bar(y=[2, 3, 1])) # or any Plotly Express function e.g. px.bar(...)
# fig.add_trace( ... )
# fig.update_layout( ... )
import dash
import dash_core_components as dcc
import dash_html_components as html
app = dash.Dash()
app.layout = html.Div([
dcc.Graph(figure=fig)
])
#app.run_server(debug=True, use_reloader=False,port=8080,) # Turn off reloader if inside Jupyter
app.run_server(debug=True, use_reloader=False,port=8080,host="0.0.0.0")

图片alt
图片alt
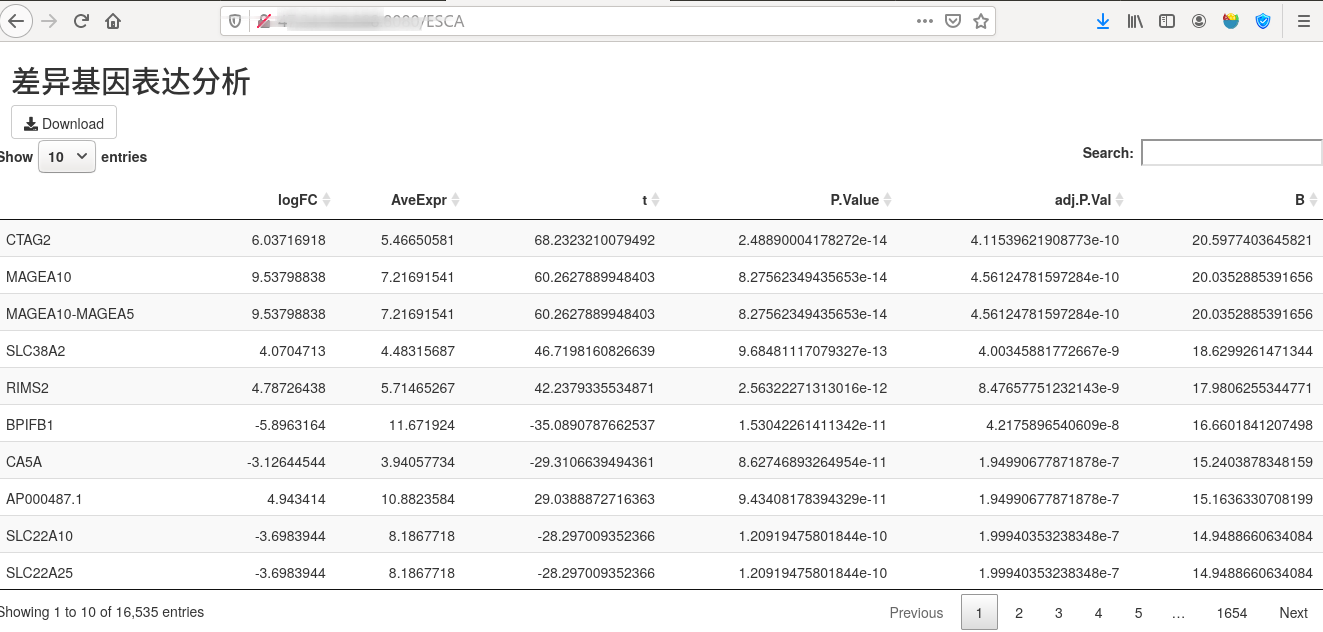
使用shiny进行数据可视化
library(shiny)
# Define UI for application that draws a histogram
ui <- fluidPage(
titlePanel("差异基因表达分析"),
downloadButton('downloadData', 'Download'),
fluidRow(
DT::dataTableOutput("table")
)
)
# Define server logic required to draw a histogram
server <- function(input, output) {
lncRNA_deg_sig <- readRDS("/home/wangyang/workspace/bioinfo_analysis/Rscript/result/limma_mRNA_deg.rda")
output$table <- DT::renderDataTable(DT::datatable({
lncRNA_deg_sig
}, rownames = T))
output$downloadData <- downloadHandler(
filename = 'file.csv',
content = function(file) {
write.csv(lncRNA_deg_sig, file)
}
)
}
# Run the application
#options(ui = ui, server = server,options=list(port=8080,host="0.0.0.0"))
shinyApp(ui = ui, server = server,uiPattern="/ESCA",options=list(port=8080,host="0.0.0.0"))
图片alt