dependence
if(F){
#sudo apt-get install libcurl4-openssl-dev
#sudo apt-get install libxml2-dev
install.packages("libxml")
install.packages("rvest")
install.packages("tidyverse")
install.packages("survivalROC")
install.packages("survival")
install.packages("survminer")
install.packages("lattice")
install.packages("ggplot2")
install.packages("ggpubr")
install.packages("devtools")
install.packages("BiocManager")
BiocManager::install("DESeq2")
BiocManager::install("TCGAbiolinks")
devtools::install_github("wangyang1749/CANCERDownload")
}
library(survivalROC)
library(survival)
library(survminer)
library(lattice)
library(DESeq2)
library(TCGAbiolinks)
library(tidyverse)
install package
devtools::install_github("wangyang1749/CANCERDownload")
数据下载
下载数据来源
- GEO
- TCGA
- UCSC xena
下载count数据
project <- "TCGA-COAD"
count <- getCount(project)
提取count数据中lncRNA和mRNA
lnRAN_mRNA_count_res <- lnRAN_mRNA(count)
lnRAN_mRNA_count_res@lnRNA
lnRAN_mRNA_count_res@mRNA
下载临床数据
clinical <- getClinical(project)
下miRNA数据
miRNACount <- getmiRNACount(project)
表达差异
DeSeq2差异基因表达分析
deg <- DeSeq2Analysis(count,project = project)
提取差异lncRNA和mRNA
lnRAN_mRNA_res <- lnRAN_mRNA(deg)
lnRAN_mRNA_res@lnRNA
lnRAN_mRNA_res@mRNA
单基因的配对差异分析
参考:单基因分析
火山图
热图
临床模型
count数据与临床数据整合
clinical_count <- clinical_count(clinical,count@count)
提取差基因
gene <- deg@deg%>%
arrange(desc(log2FoldChange))%>%
rownames_to_column("symbol")%>%
dplyr::slice(1:5)%>%
pull("symbol")
单因素Cox分析
- 可以选择差异的基因进行单因素cox分析
CoxSingle(clinical,count@count,gene)
多因素Cox分析构建风险模型
- 用单因素cox分析中有统计学意义的基因进行多因素cox分析
- 进行逐步回归
- 构建风险生存模型
coxMulti_res <- CoxMulti(clinical,count@count, gene,cutoff=Inf)
绘制ROC曲线
参考:回归分析
ROC(coxMulti_res@data,predict.time=4)
绘制生存曲线
Survival(coxMulti_res)
lasso回归构建风险模型
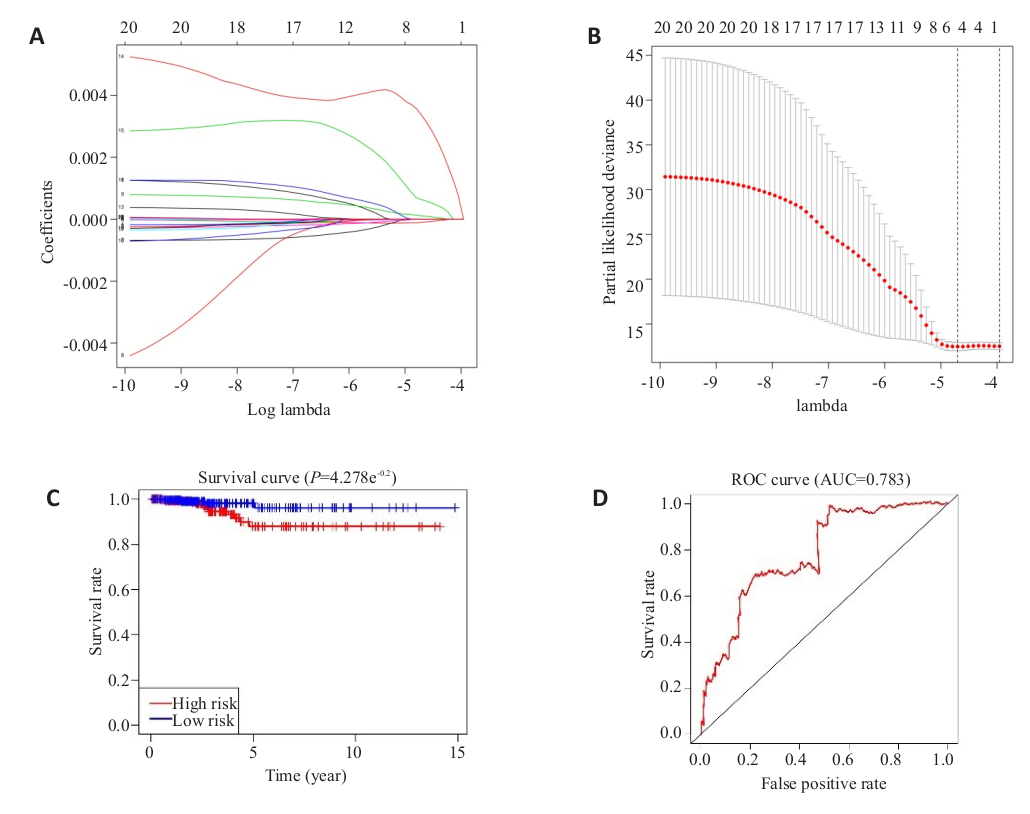
图片alt
单基因临床相关性分析
参考:单基因分析
列线图
森林图
功能注释
GO富集分析
KEGG富集分析
GSEA富集分析
分子互作
ceRNA网络
数据框的结构约定
差异表达的RNA
- circRNA_deg(circRNA difference expression gene )
- lncRNA_deg(circRNA difference expression gene )
- mRNA_deg(circRNA difference expression gene )
- miRNA_deg(circRNA difference expression gene )
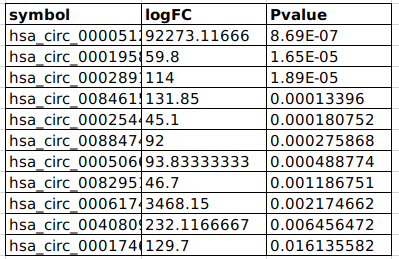
图片alt
显著的差异表达的RNA
- circRNA_deg_sig(circRNA difference expression gene Significance)
- lncRNA_deg_sig(circRNA difference expression gene Significance)
- mRNA_deg_sig(circRNA difference expression gene Significance)
- miRNA_deg_sig(circRNA difference expression gene Significance)

图片alt