S4
.hasSlot(object, name)
setOldClass("data.frame")
https://docs.tibco.com/pub/enterprise-runtime-for-R/6.0.0/doc/html/Language_Reference/methods/slot.html
创建类
setClass("Person",
slots = list(name = "character", age = "numeric"))
alice <- new("Person", name = "Alice", age = 40)
alice@name
案例
## 创建父类
setClass("Shape",
slots=list(name="character",shape="character"))
## 创建子类
setClass("Ellipse",
contains="Shape",
slots=list(radius="numeric"),
prototype=list(radius=c(1,1),shape="Ellipse"))
setClass("Circle",
contains="Ellipse",
slots=list(radius="numeric"),
prototype=list(radius = 1,shape="Circle"))
setClass("Rectangle",
contains="Shape",
slots=list(edges="numeric"),
prototype=list(edges=c(1,1),shape="Rectangle"))
setClass("Square",
contains="Rectangle",
slots=list(edges="numeric"),
prototype=list(edges=1,shape="Square"))
setGeneric("getShape",function(obj,...) {
cat("sss")
standardGeneric("getShape")
})
setMethod("getShape","Shape",function(obj,...){
cat(obj@shape,"\n")
})
setGeneric("area",function(obj,...) standardGeneric("area"))
setMethod("area","Ellipse",function(obj,...){
cat("Ellipse Area :\n")
pi * prod(obj@radius)
})
setMethod("area","Circle",function(obj,...){
cat("Circle Area :\n")
pi*obj@radius^2
})
setMethod("area","Rectangle",function(obj,...){
cat("Rectangle Area :\n")
prod(obj@edges)
})
setMethod("area","Square",function(obj,...){
cat("Square Area :\n")
obj@edges^2
})
setGeneric("circum",function(obj,...) standardGeneric("circum"))
setMethod("circum","Ellipse",function(obj,...){
cat("Ellipse Circum :\n")
2*pi*sqrt((obj@radius[1]^2+obj@radius[2]^2)/2)
})
setMethod("circum","Circle",function(obj,...){
cat("Circle Circum :\n")
2*pi*obj@radius
})
setMethod("circum","Rectangle",function(obj,...){
cat("Rectangle Circum :\n")
2*sum(obj@edges)
})
setMethod("circum","Square",function(obj,...){
cat("Square Circum :\n")
4*obj@edges
})
e1<-new("Ellipse",name="e1",radius=c(2,5))
getShape(e1)
circum(e1)
area(e1)
setValidity("Circle",function(object) {
if (object@radius <= 0) stop("Radius is negative.")
})
c1<-new("Circle",name="c1",radius=2)
getShape(c1)
circum(c1)
area(c1)
r1<-new("Rectangle",name="r1",edges=c(2,5))
getShape(r1)
circum(r1)
area(r1)
s1<-new("Square",name="s1",edges=2)
getShape(s1)
circum(s1)
area(s1)
结果
> e1<-new("Ellipse",name="e1",radius=c(2,5))
> getShape(e1)
sssEllipse
> circum(e1)
Ellipse Circum :
[1] 23.92566
> area(e1)
Ellipse Area :
[1] 31.41593

图片alt
常用方法
library(pryr)
otype(x)
getMethod("circum", "Ellipse") ## 查看泛型函数circum的Ellipse实现
S3与S4区别
S3对象:是基于泛型函数的面向对象机制.S3对象可看作是一个list并有一个名为class的属性.S3没有正式的类型定义
x <- structure(2, class = "foo")
> x<-1
> attr(x,'class')<-'foo'
> x
[1] 1
attr(,"class")
[1] "foo"
> class(x)
[1] "foo"
# 用pryr包的otype函数,检查x的类型
> otype(x)
[1] "S3"
> y <- structure(2, class = "foo")
> y
[1] 2
attr(,"class")
[1] "foo"
> class(y)
[1] "foo"
> otype(y)
[1] "S3"
S3对象的泛型和多态
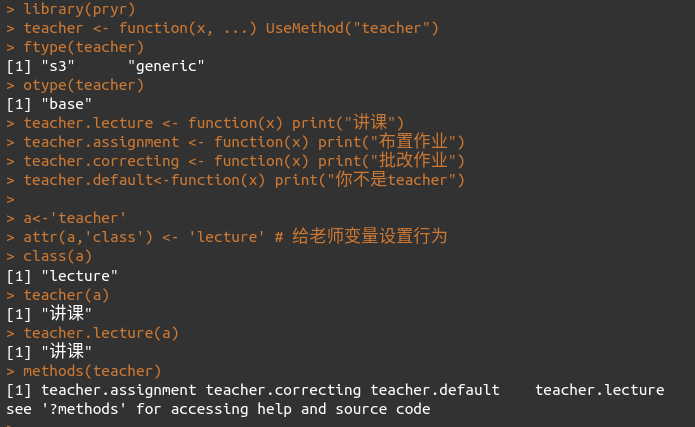
# 用UseMethod()定义teacher泛型函数
# 用teacher.xxx的语法格式定义teacher对象的行为
#其中teacher.default是默认行为
library(pryr)
teacher <- function(x, ...) UseMethod("teacher")
ftype(teacher)
otype(teacher)
teacher.lecture <- function(x) print("讲课")
teacher.assignment <- function(x) print("布置作业")
teacher.correcting <- function(x) print("批改作业")
teacher.default<-function(x) print("你不是teacher")
a<-'teacher'
attr(a,'class') <- 'lecture' # 给老师变量设置行为
class(a)
teacher(a)
teacher.lecture(a)
methods(teacher)
getAnywhere(teacher.lecture)
图片alt
plot.default(c(1,2,3))
suppressPackageStartupMessages(library(tidyverse))
library(pryr)
setClass("Shape",
slots=list(name="character",shape="character"))
# setClass("Rectangle",
# contains="Shape",
# slots=list(edges="numeric"),
# prototype=list(edges=c(1,1),shape="Rectangle"))
setGeneric("getShape",function(obj,arg1,arg2) standardGeneric("getShape"))
setMethod("getShape","Shape",function(obj,arg1,arg2) {
cat(obj@name,arg1,arg2)
})
r1<-new("Shape",name="r1")
getShape(r1,"a","sss")
otype(getShape)
ftype(getShape)
showMethods(getShape)
# 查看泛型函数函数实现
getMethod("getShape","Shape")
# > getMethod("getShape","Shape")
# Method Definition:
# function (obj, arg1, arg2)
# {
# cat(obj@name, arg1, arg2)
# }
# Signatures:
# obj
# target "Shape"
# defined "Shape"
################S3################################
getShape <- function(obj,arg1,arg2) UseMethod(generic="getShape",object=obj)
getShape.Shape <- function(obj,arg1,arg2){
cat(obj,arg1,arg2)
}
obj <- "test"
# getShape(obj) # no applicable method for 'getShape' applied to an object of class "character"
attr(obj,'class') <- 'Shape'
class(obj)
getShape(obj,"222","333")
otype(getShape)
ftype(getShape)
# 查看泛型函数函数实现
getShape.Shape
getAnywhere(getShape.Shape)
setClass("Person",slots=list(name="character",age="numeric"))
setGeneric("work",function(object) standardGeneric("work"))
setMethod("work", signature(object = "Person"), function(object) cat(object@name , "is working") )
ftype(work)
# a<-new("Person",name="Conan",age=16)
# work(a)
# ftype(work)
setClass("Shape",slots=list(name="character",shape="character"))
setGeneric("getShape",function(obj,arg1,arg2) tandardGeneric("getShape"))
setMethod("getShape","Shape",function(obj,arg1,arg2) { cat(obj@name,arg1,arg2)})
ftype(getShape)
使用$访问R S4对象
setClass("Shape",
slots=list(name="character",metadata="data.frame"))
metadata <- colData(vsd)|> as.data.frame()
#sample group sizeFactor
#IPMN-6080042 IPMN-6080042 treatment 1.84431497
#IPMN-6080180 IPMN-6080180 treatment 1.87930966
#IPMN-6100221 IPMN-6100221 treatment 0.89195100
#PDAC-1915728 PDAC-1915728 control 1.80253128
#PDAC-6050001 PDAC-6050001 control 4.35214778
#PDAC-6060065 PDAC-6060065 control 0.04433571
r1<-new("Shape",metadata=metadata)
setMethod("$", "Shape",
function(x, name)
{
cat(name)
x@metadata[[name]]
})
r1$sample
#sample[1] "IPMN-6080042" "IPMN-6080180" "IPMN-6100221" "PDAC-1915728" "PDAC-6050001" "PDAC-6060065"
修改S4对象
setClass("Shape",
slots=list(name="character",shape="character"))
setGeneric("getShape",function(obj,arg1,arg2) standardGeneric("getShape"))
setMethod("getShape","Shape",function(obj,arg1,arg2) {
cat(obj@name,arg1,arg2,"\n")
})
r1<-new("Shape",name="wangyang")
r1@name<- "sss"
getShape(r1,"a","sss")
setGeneric("init<-",function(object,value)standardGeneric("init<-"))
setMethod("init<-","Shape", function(object,value) {
object@name <- value
object
})
init(r1) <- "11111111111"
getShape(r1,"a","sss")
##################R S4 OOP的特征######################
## 泛型函数,多态的使用
## 可以函数重写(子类函数与父类同名函数)
## 没有函数重载
## 可以进行序列化与反序列化
#####################################################
#### 泛型函数
setwd("/home/wangyang/workspace/yqs136")
setClass("Shape",
slots=list(name="character",shape="character"))
setGeneric("getShape",function(obj)standardGeneric("getShape"))
setMethod("getShape","Shape",function(obj){
cat("shape: ",obj@shape," name: ",obj@name,"\n")
})
setClass("Rectangle",
contains="Shape",
slots=list(edges="numeric"),
prototype=list(shape="Rectangle"))
setClass("Circle",
contains="Shape",
slots=list(radius="numeric"),
prototype=list(shape="Circle"))
r1<-new("Rectangle",name="Rectangle001",edges=c(2,5))
c2<-new("Circle",name="Circle002",radius=2)
slot(r1,"name")
r1@name
getShape(r1)
getShape(c2)
#### 多态 注:R可以函数重写,就是子类函数可以覆盖父类的同名函数
setMethod("getShape","Rectangle",function(obj){
cat("Rectangle的getShape函数已经被重写!\n")
})
getShape(r1)
getShape(c2)
setMethod("getShape","Circle",function(obj){
cat("Circle的getShape函数已经被重写!\n")
})
getShape(r1)
getShape(c2)
## 没有函数重载
setGeneric("area",function(obj) standardGeneric("area"))
setMethod("area","Circle",function(obj){
cat("Circle Area :\n")
pi*obj@radius^2
})
setMethod("area","Rectangle",function(obj){
cat("Rectangle Area :\n")
prod(obj@edges)
})
area(r1)
area(c2)
setGeneric("area",function(obj,value) standardGeneric("area"))
setMethod("area","Shape",function(obj,value){
cat(obj@name,value,"\n")
})
area(r1,"aa")
area(r1,"bb")
## 可以进行序列化与反序列化
saveRDS(r1,file="rds/r1.rds")
r1 <- readRDS("rds/r1.rds")
r1@name
##################使用 R S4带来的好处#####################
# 将结果存储在一个对象里
# 快速修改函数的实现
# 函数debug方式1:给参数附上具体的值
# 函数debug方式2:通过Debug的方式进行调试
########################################################
#### 将结果存储在一个对象里
source("/home/wangyang/workspace/yqs136/R/S4/class.R")
miRNA_Obj <- readRDS("/home/wangyang/workspace/yqs136/rds/miRNA_Obj.rds")
piRNA_Obj <- readRDS("/home/wangyang/workspace/yqs136/rds/piRNA_Obj.rds")
miRNA_Obj@DegDFs[["miRNA_4023_vs_2558_RWP1"]]@deg |> write_tsv(file="deg/miRNA_4023_vs_2558_RWP1.tsv")
miRNA_Obj@DegDFs[["miRNA_IPMN_vs_PDAC"]]@deg |> write_tsv(file="deg/miRNA_IPMN_vs_PDAC.tsv")
miRNA_Obj@DegDFs[["miRNA_NORMAL_vs_IPMN"]]@deg |> write_tsv(file="deg/miRNA_NORMAL_vs_IPMN.tsv")
miRNA_Obj@DegDFs[["miRNA_SCRAMBLE_vs_2558_RWP1"]]@deg |> write_tsv(file="deg/miRNA_SCRAMBLE_vs_2558_RWP1.tsv")
piRNA_Obj@DegDFs[["piRNA_4023_vs_2558_RWP1"]]@deg |> write_tsv(file="deg/piRNA_4023_vs_2558_RWP1.tsv")
piRNA_Obj@DegDFs[["piRNA_IPMN_vs_PDAC"]]@deg |> write_tsv(file="deg/piRNA_IPMN_vs_PDACtsv")
piRNA_Obj@DegDFs[["piRNA_NORMAL_vs_IPMN"]]@deg |> write_tsv(file="deg/piRNA_NORMAL_vs_IPMN.tsv")
piRNA_Obj@DegDFs[["piRNA_SCRAMBLE_vs_2558_RWP1"]]@deg |> write_tsv(file="deg/piRNA_SCRAMBLE_vs_2558_RWP1.tsv")
miRNA_Obj <- volcano(miRNA_Obj,name="miRNA_4023_vs_2558_RWP1")
miRNA_Obj <- intersectGene(miRNA_Obj)
# 查看miRNA_4023_vs_2558_RWP1组差异基因个数
miRNA_Obj@DegDFs[["miRNA_4023_vs_2558_RWP1"]]@deg |> dim()
# 查看miRNA_4023_vs_2558_RWP1组数据上调和下调差异基因
table(miRNA_Obj@DegDFs[["miRNA_4023_vs_2558_RWP1"]]@degSig$direction)
# 查看miRNA_4023_vs_2558_RWP1组所有差异的基因
miRNA_Obj@DegDFs[["miRNA_4023_vs_2558_RWP1"]]@degSig$symbol
getwd()
#### 快速修改函数的实现
source("/home/wangyang/workspace/yqs136/R/S4/class.R")
miRNA_Obj <- volcano(miRNA_Obj,name="miRNA_SCRAMBLE_vs_2558_RWP1")
miRNA_Obj@DegDFs[["miRNA_4023_vs_2558_RWP1"]]@expr
setMethod("runDeg","YQS136",function(obj,name,...){
metadata <- getMetadata(obj,...)
expr <- obj@expr[,metadata$sample]
metadata$group <- factor(metadata$group,levels=c("control","treatment"))
metadata$batch <- factor(metadata$batch)
message("expr column and metadata row: ",identical(colnames(expr),metadata$sample))
####################limma的逻辑###########################
{
design <- model.matrix(~metadata$group)
dge <- DGEList(counts=expr)
keep <- filterByExpr(dge, design)
dge <- dge[keep,,keep.lib.sizes=FALSE]
dge <- calcNormFactors(dge)
logCPM <- cpm(dge, log=TRUE, prior.count=3)
fit <- lmFit(logCPM, design)
fit <- eBayes(fit, trend=TRUE)
deg <- topTreat(fit, coef=ncol(design),n = Inf) |>
rownames_to_column("symbol")|>
select(symbol,log2FoldChange=logFC,pvalue=P.Value,padj=adj.P.Val)
}
### deg结果的命名规范必须是如下,以提供给volcano和heatmap函数使用
#symbol baseMean log2FoldChange lfcSE stat pvalue padj
#hsa-let-7a 1.444046e+03 0.0902414404 0.2895802 0.3116284873 0.75532288 0.9997432
##################limma的逻辑###########################
if(name %in% names(obj@DegDFs)){
obj@DegDFs[[name]] <- new("DegDF",expr=expr,metadata=metadata,deg=deg)
# obj@DegDFs <- c(obj@DegDFs,DegDF_list)
}else{
DegDF_list <- list(new("DegDF",expr=expr,metadata=metadata,deg=deg))
names(DegDF_list) <- name
obj@DegDFs <- c(obj@DegDFs,DegDF_list)
}
obj
})
miRNA_Obj <-runDeg(miRNA_Obj,
name="miRNA_SCRAMBLE_vs_2558_RWP1",
treatment=c("2558-SCRAMBLE","RWP1-SCRAMBLE"),
control=c(miRNA_Obj@group$cell_2558,miRNA_Obj@group$cell_RWP1),
treatment_name="SCRAMBLE",
control_name="2558&RWP1")
miRNA_Obj <- volcano(miRNA_Obj,name="miRNA_SCRAMBLE_vs_2558_RWP1")
miRNA_Obj <- intersectGene(miRNA_Obj)
setMethod("intersectGene","YQS136",function(obj){
gene_list <- lapply(1:length(obj@DegDFs),function(x){obj@DegDFs[[x]]@degSig$symbol})
names(gene_list) <- names(obj@DegDFs)
gene_list <- gene_list[lengths(gene_list) != 0]
obj@intersectGene <- Reduce(function(x,y) intersect(x,y), gene_list,accumulate =FALSE)
{
myCol <- brewer.pal(length(gene_list), "Set1")
res <- venn(gene_list, zcolor = myCol,box=F,opacity = 0.3)
#ggvenn( gene_list,stroke_size = 0.5, set_name_size = 3, text_size = 2)
}
obj
})
miRNA_Obj <- intersectGene(miRNA_Obj)
#### 函数调试方式1:给参数附上具体的值
metadata <- getMetadata(miRNA_Obj,
treatment=c("2558-SCRAMBLE","RWP1-SCRAMBLE"),
control=c(miRNA_Obj@group$cell_2558,miRNA_Obj@group$cell_RWP1),
treatment_name="SCRAMBLE",
control_name="2558&RWP1")
name <- "miRNA_SCRAMBLE_vs_2558_RWP1"
obj <- miRNA_Obj
#### 函数debug方式2:通过Debug的方式进行调试
#' Print method for "foo" class
#'
#' @param x An object of class "foo"
#' @param ... Other arguments passed to or from other methods
#'
#' @export print.foo
#' @export
print.foo <- function(x, ...) {
cat("This is just a dummy function.\n")
}
# Generated by roxygen2: do not edit by hand
S3method(print,foo)
export(print.foo)
https://stackoverflow.com/questions/18512528/how-to-export-s3-method-so-it-is-available-in-namespace
图片alt
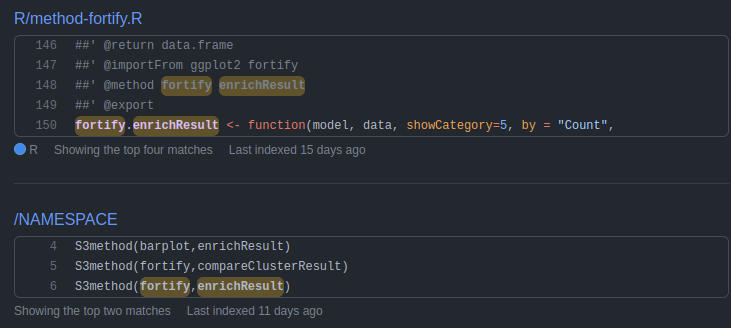
https://github.com/YuLab-SMU/DOSE/blob/master/R/accessor.R#L84
##' @method [[ enrichResult
##' @export
`[[.enrichResult` <- function(x, i) {
gc <- geneInCategory(x)
if (!i %in% names(gc))
stop("input term not found...")
gc[[i]]
}
stampedEnv <- setClass("stampedEnv", contains = "environment",
slots = c(update = "POSIXct"))
setMethod("[[<-", c("stampedEnv", "character", "missing"),
function(x, i, j, ..., value) {
ev <- as(x, "environment")
ev[[i]] <- value #update the object in the environment
x@update <- Sys.time() # and the update time
x})
e1 <- stampedEnv(update = Sys.time())
e1[["noise"]] <- rnorm(10)