1 相关分析
1.1 计算相关系数

图片alt
library(corrplot)
library(tidyverse)
M <- cor(mtcars)
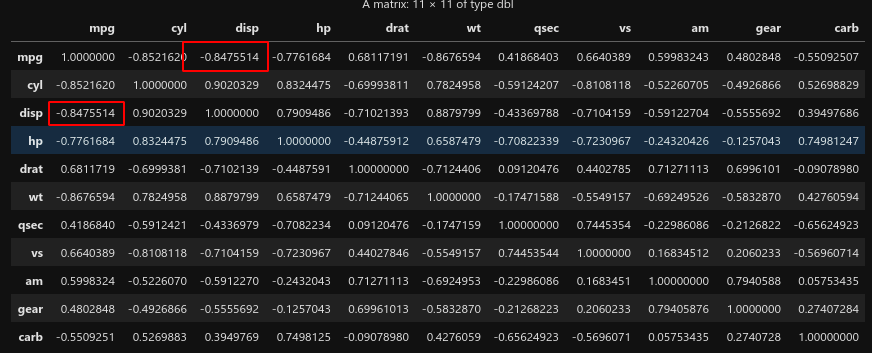
图片alt
attach(mtcars)
a <- sum( (mpg - mean(mpg)) * (disp-mean(disp)) )
b <- sqrt( sum( (mpg-mean(mpg))^2 ) * sum( (disp-mean(disp))^2 ) )
r <- a/b
r
cor(mpg,disp)
#cor.mtest(count,conf.level=0.95)
detach(mtcars)
-0.847551379262479
-0.847551379262479
1.2 相关系数假设检验
r /sqrt( (1-r^2)/( length(mpg) -2 ) )
cor.test(mpg,disp)
-8.74715153409391
Pearson's product-moment correlation
data: mpg and disp
t = -8.7472, df = 30, p-value = 9.38e-10
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
-0.9233594 -0.7081376
sample estimates:
cor
-0.8475514
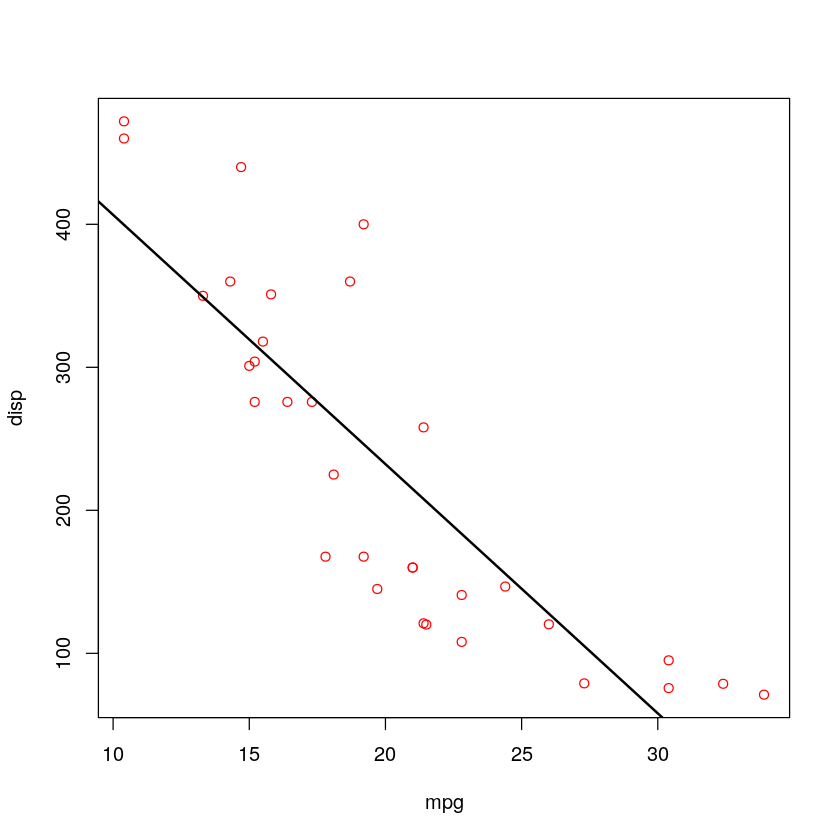
由于p=9.38e-10<0.05, 于是在\alpha =0.05的水准上拒绝H_0, 接受H_1, 可认为mpg与disp呈现负的线性相关。
with(mtcars,plot(mpg,disp,type="n"))
with(mtcars,points(mpg,disp,col="red"))
fit = lm(disp~mpg,mtcars)
abline(fit,lwd=2)
图片alt
keep <- rowSums(expr_lnRNA > 0.5)>=2
expr_lnRNA <- expr_lnRNA[keep,]
#lnRNA_count<-lnRNA_count[!apply(lnRNA_count, 1,mean)==0,]
cor_result <- (function(m6a_count,lnRNA_count){
cor_result <- data.frame()
for (mRNA in rownames(m6a_count)){
#exp_cor<-rbind(m6a_count[1,],lnRNA_count)
y <- as.numeric(m6a_count[mRNA,])
for (lnRNA in rownames(lnRNA_count)){
x <- as.numeric(lnRNA_count[lnRNA,])
test <- cor.test(x,y,type="pearson")
cor_result <- rbind(mRNA=mRNA,lnRNA=lnRNA,cor_result,data.frame(pvalue=test$p.value,estimate=test$estimate))
}
}
return(cor_result)
})(lnRAN_mRNA(fpkm_obj)@mRNA[gene,],expr_lnRNA)
WGCNA
回归流程
- 单因素cox分析找到预后相关基因
- 多因素cox分析构建临床风险模型
- 以风险中位数将病人分为高风险组和低风险组
- 构建风险生存曲线
- 风险与临床相关性检验
- 风险与临床相关性热图
多因素逐步回归
单因素cox回归

图片alt
多因素cox回归
lasso回归
图形展示
森林图
多因素回归分析时,为探讨影响因素对结局事件的影响大小,可以利用森林图更直观的将回归结果可视化

图片alt
列线图(Nomogram)
使用Nomogram对结局事件的发生风险进行预测,列线图将复杂的回归方程,转变为了可视化的图形,使预测模型的结果更具有可读性
library(survival)
library(rms)
dd<-datadist(lung)
options(datadist="dd")
f <- cph(Surv(time,status)~age+sex,data=lung,x=TRUE,y=TRUE,surv=TRUE)
survival <- Survival(f)
survival1 <- function(x)survival(365,x)
survival2 <- function(x)survival(730,x)
nom <- nomogram(f,fun = list(survival1,survival2),
fun.at = c(0.05,seq(0.1,0.9,by=0.05),0.95))
plot(nom)
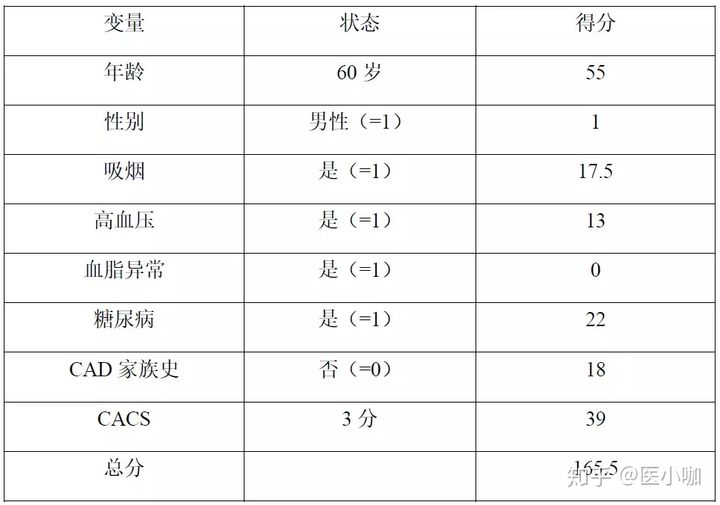
- 预测患者对应的未来5年、10年和15年的生存率

图片alt
图片alt